About
|
Background
In general, microarray-based and next generation sequencing (NGS)-based technologies are the two major resources of gene expression datasets. The improvement of high-throughput sequencing facilitates the transcriptome analysis. the threshold of transcriptome sequencing is reduced. Hence, NGS has become the mainstream for monitoring gene expression levels in parallel with various experimental treatments. Unfortunately, most researchers often have no idea to perform further analysis based on the preliminary data after finishing the process of annotation. |
|
Description
EXPath Tool was developed for the pathway annotation, display, and comparative analysis of user-customized gene expression profiles derived from microarray platforms or next-generation sequencing (NGS) technology under various conditions to infer metabolic pathways for all organisms. By using EXPath Tool, user can create their own project and import the annotation files derived from either microarray or NGS platform. After the step of data preprocessing is complete, user can view the project and: 1. Access the gene expression patterns and the candidates of co-expression genes (Co-expression analysis). 2. Dissect differentially expressed genes (DEGs) between two conditions (DEGs search), functional grouping with pathway and GO (Pathway / GO enrichment analysis), and correlation networks (Co-expression analysis). 3. View the expression patterns of genes involved in specific pathways to infer the effect of the treatment. |
Conclusions
In conclusion, EXPath tool, freely available at http://expathtool.itps.ncku.edu.tw/, is a web-based platform that integrates gene expression data with metabolic pathways. We believe that EXPath tool may provide an effective way for users to explore the inferred pathway information that will be valuable for further research. |
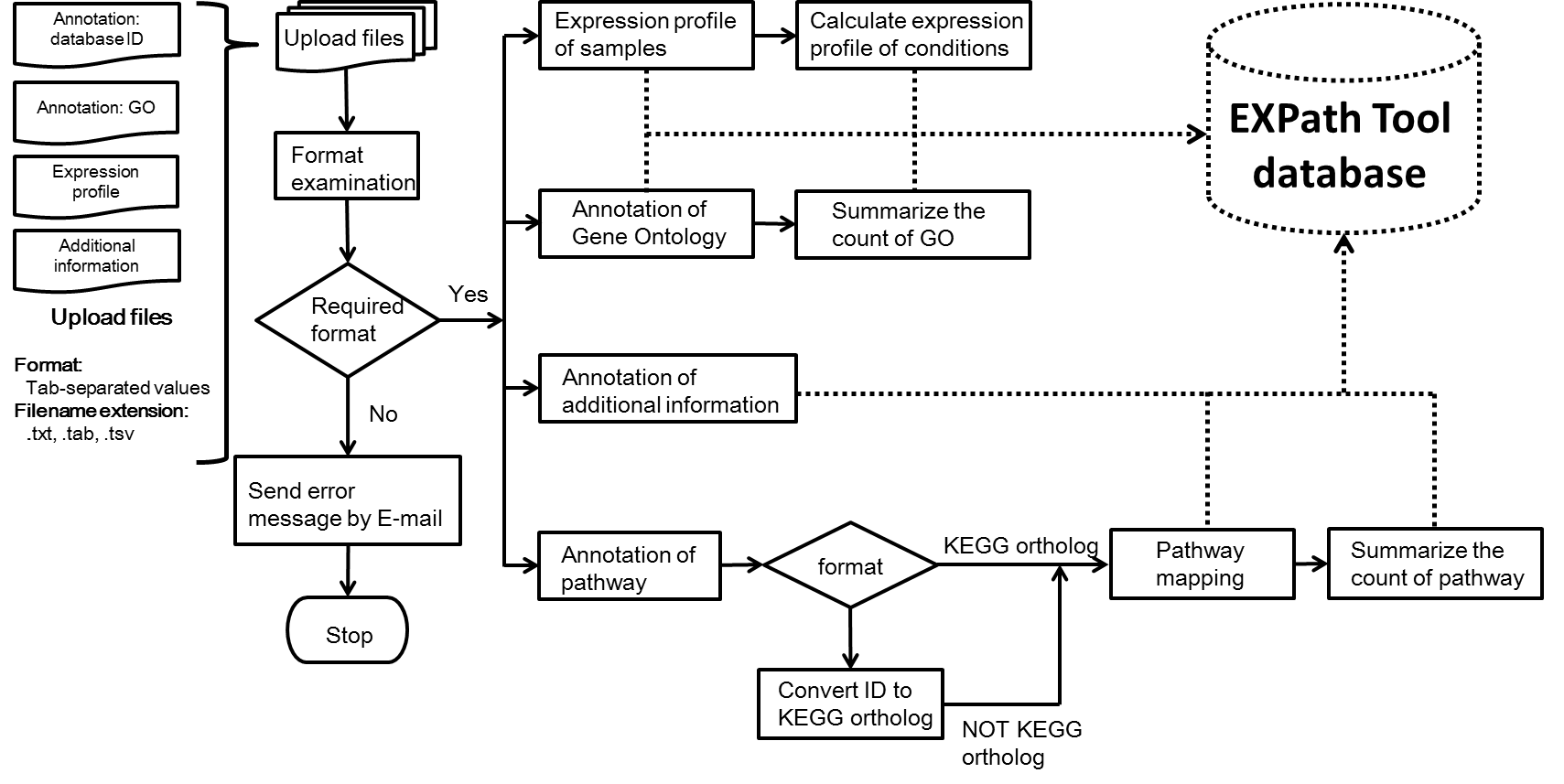 Figure 1. The analysis flowchart of data processing in EXPath Tool system.
Figure 1. The analysis flowchart of data processing in EXPath Tool system.
|
